antiSMASH 8.0: The leading tool for detecting and characterising biosynthetic gene clusters in bacteria and fungi
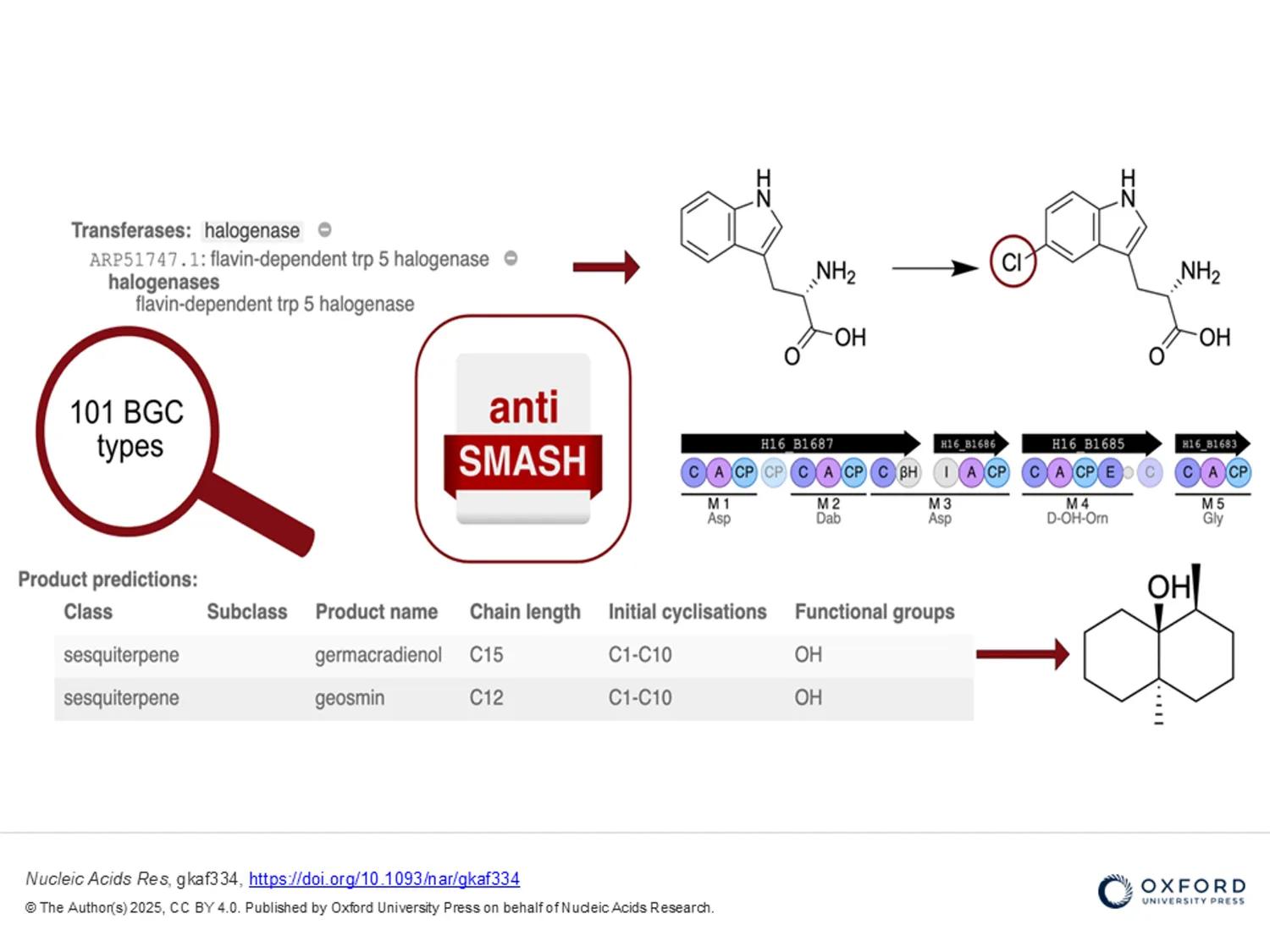
Since its inception in 2011, antiSMASH has become the leading tool for detecting and characterising biosynthetic gene clusters (BGCs) in microbes such as bacteria and fungi. A BGC can be defined as a physically clustered group of two or more genes in a particular genome that together encode a biosynthetic pathway for the production of a specialised metabolite, including its chemical variants (Medema et al. 2015). Apart from playing an essential role in microbial interactions, these metabolites are also crucial for the development of pharmaceuticals and agrochemicals. The discovery of several BGCs directly from microbial genomes has been made possible by developments in genome sequencing in recent decades.
This new publication involving MARBLES partners University of Leiden (ULEI) and Wageningen University (WU), presents the latest newly developed antiSMASH version. antiSMASH 8.0 has improved capabilities for analysing BGCs, chemistry, enzymes and regulation. It has increased the number of detectable cluster types from 81 to 101, has improved analysis support for terpenoids and tailoring enzymes, and improved the analysis of modular enzymes like polyketide synthases and nonribosomal peptide synthetases. These enhancements expand antiSMASH's overall predictive capabilities for natural product genome mining, ensuring it's up-to-date with industry advancements.
Blin, K., Shaw, S., Vader, L., Szenei, J., Reitz, Z. L., Augustijn, H. E., Cediel-Becerra, J. D. D., De Crécy-Lagard, V., Koetsier, R. A., Williams, S. E., Cruz-Morales, P., Wongwas, S., Luchsinger, A. E. S., Biermann, F., Korenskaia, A., Zdouc, M. M., Meijer, D., Terlouw, B. R., Van Der Hooft, J. J. J., Ziemert, N., Helfrich, E.J.N., Masschelein, J., Corre, C., Chevrette, M.G., van Wezel, G.P., Medema, M.H. & Weber, T. (2025). antiSMASH 8.0: extended gene cluster detection capabilities and analyses of chemistry, enzymology, and regulation. Nucleic Acids Research. DOI: https://doi.org/10.1093/nar/gkaf334
Medema, M. H., Kottmann, R., Yilmaz, P., Cummings, M., Biggins, J. B., Blin, K., De Bruijn, I., Chooi, Y. H., Claesen, J., Coates, R. C., Cruz-Morales, P., Duddela, S., Düsterhus, S., Edwards, D. J., Fewer, D. P., Garg, N., Geiger, C., Gomez-Escribano, J. P., Greule, A., . . . Glöckner, F. O. (2015). Minimum Information about a Biosynthetic Gene cluster. Nature Chemical Biology, 11(9), 625–631. https://doi.org/10.1038/nchembio.1890