A smarter way to search for antibiotics
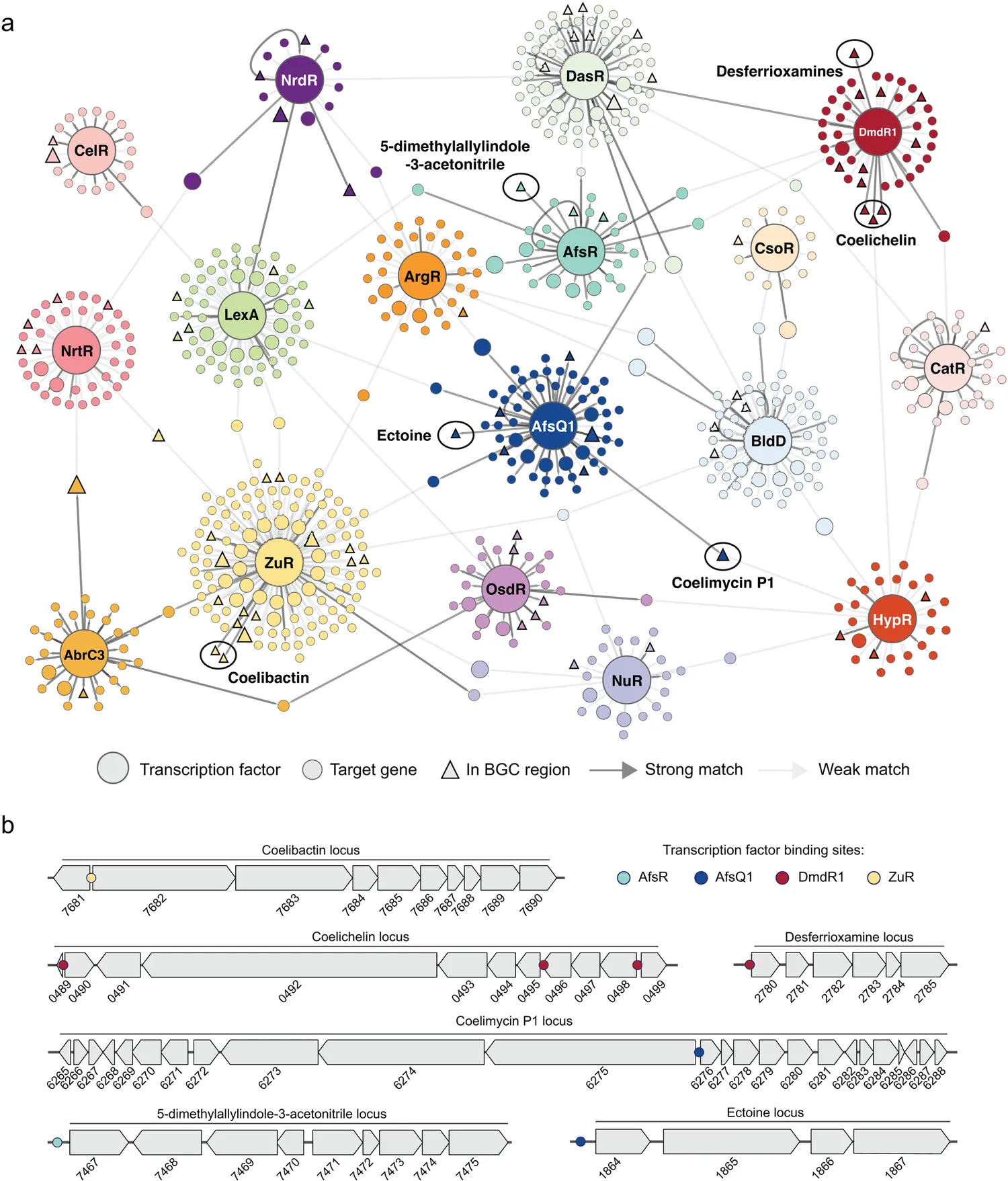
A key aspect of the European Union ERC Advanced Grant-funded COMMUNITY project was to predict biosynthetic gene cluster (BGC) function entirely on how BGCs are controlled, without looking at predicted gene function or natural products. Gilles van Wezel, University of Leiden (MARBLES coordinator), and his team within the COMMUNITY project have developed a new and more effective way to search for antibiotics. They focus on when certain bits of DNA are switched on or off. You can predict what a piece of DNA will do if you can locate the "switch" that controls it, e.g., whether it plays a role in producing an antibiotic. In bacteria such as Streptomyces, there are still millions of BGCs whose potential is unknown. The team expects that this new approach will help them find the most valuable individuals faster.
In the newly published research, they have shown that control by the iron master regulator in Streptomyces bacteria can be used as a beacon to find new genes for the biosynthesis of the siderophore desferrioxamine.
Read the open access publication:
Augustijn, H. E., Reitz, Z. L., Zhang, L., Boot, J. A., Elsayed, S. S., Challis, G. L., Medema, M. H., & van Wezel, G. P. (2025). Genome mining based on transcriptional regulatory networks uncovers a novel locus involved in desferrioxamine biosynthesis. PLOS Biology, 23(6). https://doi.org/10.1371/journal.pbio.3003183